※ Links:
Introduction:
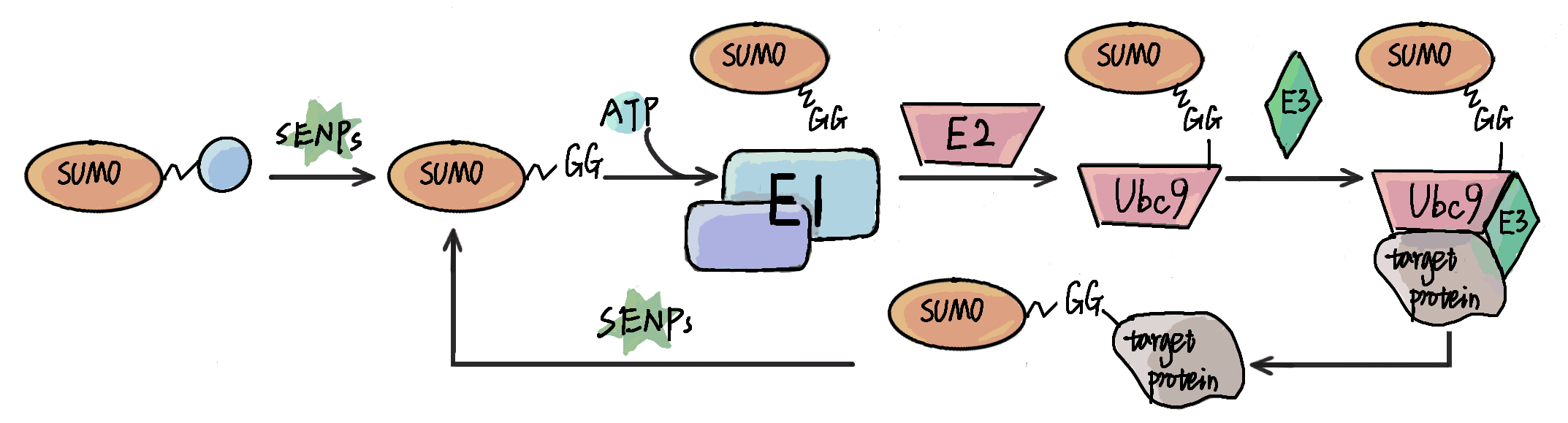
Protein SUMOylation is one of the most ubiquitous post-translational modification (PTM), and plays important roles in most of biological processes. Besides experimental approaches, prediction of potential candidates with computational methods has also attracted great attention for its convenience and fast-speed. In this review, we present a comprehensive but brief summarization of computational resources of protein SUMOylation, including SUMOylation databases, prediction of SUMOylation sites and other tools.
We appreciate all help from databases and tools therefore we present references about them. We are grateful for users feedback. Please inform Dr. Yu Xue, Dr. Di Peng, Yujie Gou or Dan Liu to add, remove or update one or multiple web links below.
Index:
<2> 3D Sturcture Databases
<3> PPI Sturcture Databases
<4> Implemented Tools
<5> Predictor of SUMOylation Sites
<6> Miscellaneous Tools
==================================================================================
1. dbPTM: A comprehensive database for experimentally identified PTMs with annotations (Huang, et al., 2019).
2. ProteomeScount: An online database for PTMs in proteins (Matlock, et al., 2015).
3. iPTMnet: An integrated database of protein PTMs in the context of systems biology. (Huang, et al., 2018).
4. BioGRID: A public database that curated genetic, chemical interactions and proteins with a large number of PTM sites (Oughtred, et al., 2019).
5. PhosphoSitePlus: A comprehensive and well annotated resource of multiple PTMs in proteins (Hornbeck, et al., 2019).
6. mUbiSiDa: A resource contains experimentally validated ubiquitinated proteins with sites for several species (Chen, et al., 2014).
7. HPRD: The human protein reference database that contains a lot of annotations including PTMs (Keshava, et al., 2009).
8. ActiveDriverDB: An online database developed to visualize and explore mutations affecting PTM sites in human proteins/genes (Krassowski, et al., 2018).
9. UniProt: The universal protein knowledgebase contains lots of protein modification information including PTMs (UniProt Consortium, et al., 2021).
10. CPLM: CPLM 4.0 contained 592,606 modification events in 463,156 unique sites of 105,673 proteins for up to 29 PLM types across 219 species (Zhang, et al., 2021).
11. qPTM: A new version of qPhos database, is a web-based database for 6 types of PTMs including acetylation, glycosylation, methylation, phosphorylation, SUMOylation, ubiquitylation in 4 different organisms including human, mouse, rat and yeast. Also, the matched proteome datasets were integrated if available. In total, 11,482,533 quantification events for 660,030 sites on 40,728 proteins under 2,596 conditions are collected and integrated in the qPTM database (Yu K, et al., 2022).
12. VPTMdb: The first comprehensive viral posttranslational modification database (VPTMdb) for collecting systematic information of PTMs in human viruses and infected host cells (Xiang, et al., 2021).
13. PTMcode v2: A database covers 19 eukaryotic species from which we collected more than 300,000 experimentally verified PTMs (>1,300,000 propagated) of 69 types extracting the post-translational regulation of >100,000 proteins and >100,000 interactions (Minguez, et al., 2015).
1. PDB: A leading resource of structural data of biological macromolecules (Berman, et al., 2000).
2. MMDB: A database links protein 3D structure data, the sequence data and classification resources with PubChem (Madej, et al., 2012).
3. SCOP: A database provides the framework for protein structure classification and annotation (Andreeva, et al., 2014).
4. DNAproDB: A database contains biochemical features and structural from structures of DNA-protein complexes (Sagendorf, et al., 2020).
5. MobiDB 3.0: A database of protein disorder and mobility annotations (Piovesan, et al., 2018).
1. iRefIndex: An integrative resource of PPIs (Razick, et al., 2008).
2. Mentha: A well curated PPI database (Anwar, et al., 2019).
3. DifferentialNet: A novel database that provides differential protein-protein networks of human tissues (Basha, et al., 2018).
4. TIMBAL: A database containing protein-protein interactions and molecules that modulate PPIs (Higueruelo, et al., 2013).
5. MIST: A helpful resource for annotating gene-protein and protein-protein interactions (Hu, et al., 2018).
6. IID: A well curated PPI database (Kotlyar, et al., 2016).
7. HIPPIE: An integrated database of protein-protein interactions. (Alanis-Lobato, et al., 2017).
8. PINA: A well curated PPI database (Du, et al., 2021).
9. HINT: A curated compilation of high-quality protein-protein interactions (Jishnu, et al., 2012).
10. inBio MapTM : A scored human protein-protein interaction network database (Zhen, et al., 2018).
11. STRING: A database of known and predicted protein-protein interactions, covers 9,643,763 proteins from 2,031 organisms (Damian, et al., 2019).
12. HPRD: The human protein reference database that contains a lot of annotations including PPIs (Keshava, et al., 2009).
13. BioGRID: A public database that curated genetic, chemical interactions and proteins with a large number of PPIs (Oughtred, et al., 2019).
1. Apache ECharts: An Open Source JavaScript Visualization Library.
2. IUPred: The web server takes a single amino acid sequence as an input and calculates the pairwise energy profile along the sequence (Erdos, et al., 2021).
3. 3Dmol.js: A modern, object-oriented JavaScript library for visualizing molecular data.
4. NetSurfP: A tool for predicting solvent accessibility, secondary structure, structural disorder and backbone dihedral angles for each residue of an amino acid sequence (Hoie, et al., 2022).
<5> Predictor of SUMOylation sites:
1. MusiteDeep : A deep-learning based webserver for protein post-translational modification site prediction and visualization (Wang, et al., 2020).
2. GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs (Zhao, et al., 2014).
3. JASSA: A comprehensive tool for prediction of SUMOylation sites and SIMs (Beauclair, et al., 2015).
4. iAcet-Sumo: Identification of lysine acetylation and sumoylation sites in proteins by multi-class transformation methods (Yang, et al., 2018).
5. SUMOplot: The presence of this post-translational modification may help explain larger MWs than expected on SDS gels due to attachment of SUMO protein at multiple positions in your protein.
6. SUMOsp 2.0: Development of a site-specific predictor of SUMOsp 2.0 ( Ren, et al., 2009).
1. GPS
6.0 ![]() : An update on the prediction of kinase-specific phosphorylation sites in proteins
(Chen, et al., 2023).
: An update on the prediction of kinase-specific phosphorylation sites in proteins
(Chen, et al., 2023).
2. HemI
2.0 ![]() : An online service for heatmap illustration
(Ning, et al., 2022).
: An online service for heatmap illustration
(Ning, et al., 2022).