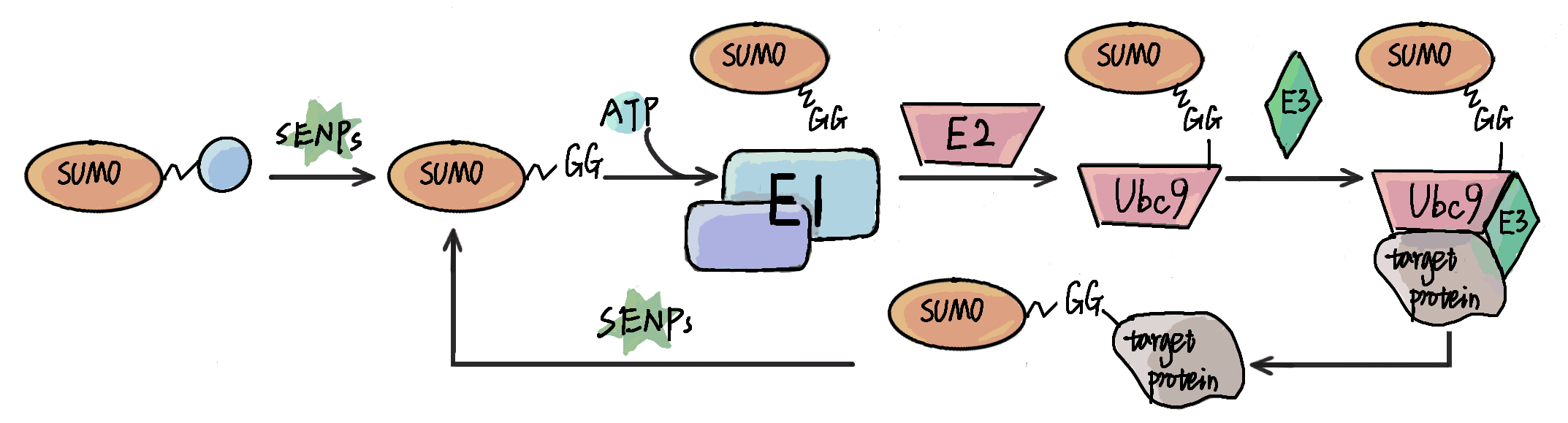
※ Advanced Services of GPS-SUMO 2.0 
Please click the check box '![]() ' to select your predictor, you can see '
' to select your predictor, you can see '![]() ' before the selected one.
' before the selected one.
![]() 1. GPS-SUMO 2.0 (PLR + GSEA): Prediction based on penalized logistic regression + GSEA with the group-based prediction system feature (Also available at the HOME page).
1. GPS-SUMO 2.0 (PLR + GSEA): Prediction based on penalized logistic regression + GSEA with the group-based prediction system feature (Also available at the HOME page). ![]() Speed:
Speed: ![]()
![]()
![]()
![]()
![]()
![]() 2. GPS-SUMO 2.0 (Transformer): Prediction based on Transformer with the contextual information.
2. GPS-SUMO 2.0 (Transformer): Prediction based on Transformer with the contextual information. ![]() Speed:
Speed: ![]()
![]()
![]()
![]()
![]() 3. GPS-SUMO 2.0 (All): Prediction based on all models with all features.
3. GPS-SUMO 2.0 (All): Prediction based on all models with all features. ![]() Speed:
Speed: ![]()
![]()
![]()
![]() 4. GPS-SUMO 2.0 (Species-specific): Species-specific prediction based on all models with all features.
4. GPS-SUMO 2.0 (Species-specific): Species-specific prediction based on all models with all features. ![]() Speed:
Speed: ![]()
![]()
![]() 5. GPS-SUMO 2.0 (Comprehensive): Prediction based on all models with all features and additional annotations of secondary structure and surface accessibility.
5. GPS-SUMO 2.0 (Comprehensive): Prediction based on all models with all features and additional annotations of secondary structure and surface accessibility. ![]() Speed:
Speed: ![]()
![]() 6. GPS-SUMO 2.0 (Stress conditions): Prediction based on penalized logistic regression, using 39,938 non-redundant SUMOylation sites identified under various stress conditions, such as SUMO protease inhibition, proteasome inhibition and heat shock.
6. GPS-SUMO 2.0 (Stress conditions): Prediction based on penalized logistic regression, using 39,938 non-redundant SUMOylation sites identified under various stress conditions, such as SUMO protease inhibition, proteasome inhibition and heat shock. ![]() Speed:
Speed: ![]()
![]()
![]()
![]()
![]()
![]() According to
According to
![]() Sequence(s) in FASTA format
Sequence(s) in FASTA format ![]() UniProt Accession Number(s)
UniProt Accession Number(s)
Please enter sequence(s) in FASTA format
Or, upload file (<50K) Unselected
You could input one
primary sequence or
multiple proteins' sequences
in FASTA format which begins with '>'.
And please don't input any special characters.
After GPS-SUMO 2.0 predictor model was well-trained, we performed an evaluation on this model. From the evaluation, three thresholds with High, Medium and Low stringency were chosen for GPS-SUMO 2.0. The performance under these three thresholds was presented as follow:
| Threshold | SUMOylation | SUMO interaction | ||||||||
| Ac | Sn | Sp | MCC | Pr | Ac | Sn | Sp | MCC | Pr | |
| High | 88.63% | 57.45% | 95.17% | 0.5749 | 71.39% | 94.64% | 90.06% | 95.17% | 0.7551 | 68.08% |
| Medium | 86.60% | 68.24% | 90.46% | 0.5585 | 60.00% | 90.88% | 98.14% | 90.05% | 0.6823 | 53.02% |
| Low | 84.33% | 75.98% | 85.01% | 0.5293 | 51.54% | 86.80% | 99.38% | 85.36% | 0.6081 | 43.72% |